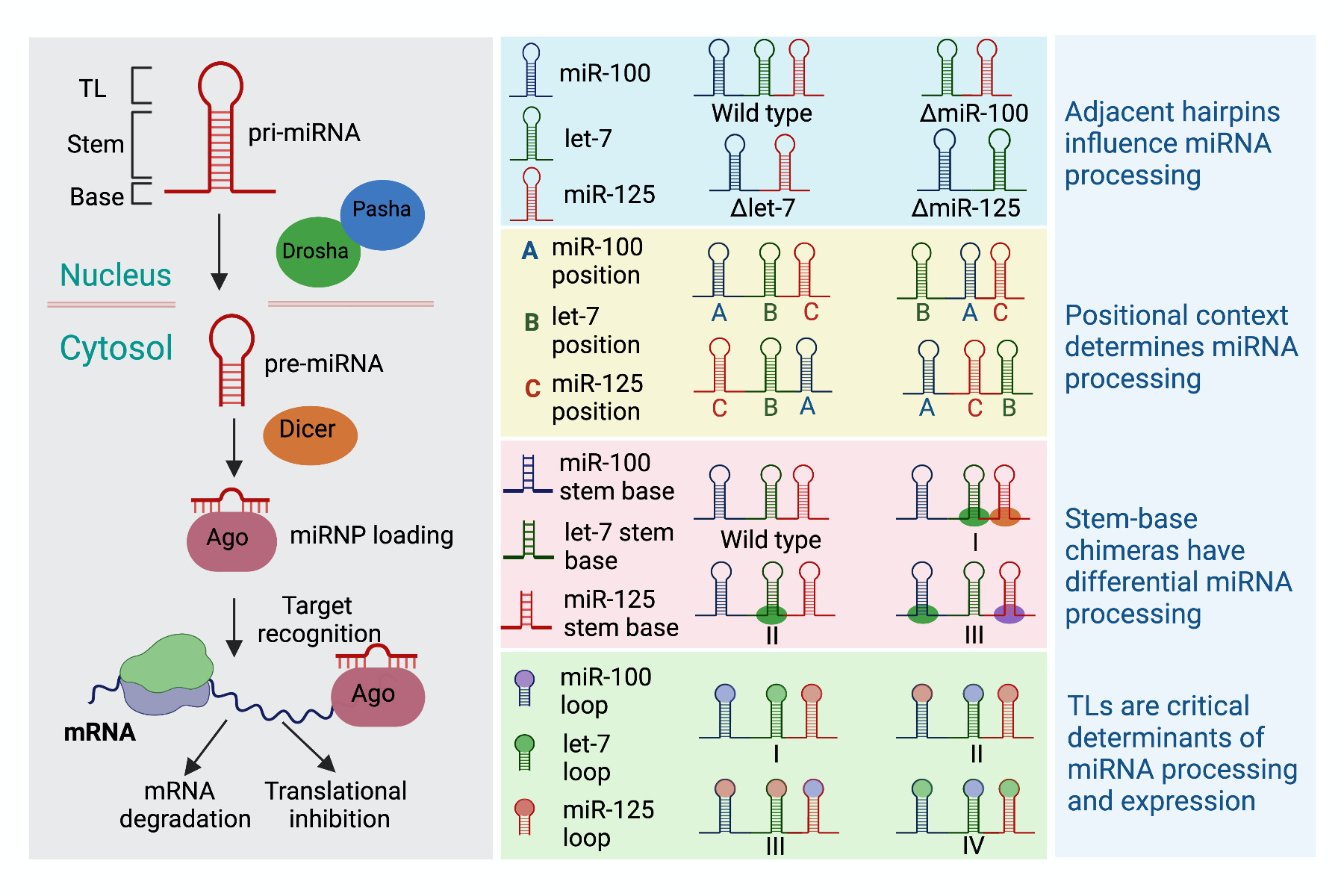
 MicroRNAs (miRNAs) belong to the class of small noncoding RNAs that regulate gene expression at the post-transcriptional level. Approximately 50% of miRNA loci in Drosophila melanogaster reside in close proximity in the genome to form clusters that are transcribed together. Since, these clusters are transcribed as a single primary transcript, fine-tuning of the downstream effector pathways is achieved by post-transcriptional regulatory mechanisms that determine the processing efficiency. The cis-acting sequence and structural features of the primary miRNA that interact with the processing machinery are critical determinants of the processing efficiency of the primary and precursor miRNAs. To elucidate the structural attributes of a polycistronic transcript that contribute towards the differences in efficiencies of processing of the co-transcribed miRNAs, we constructed a series of chimeric variants of Drosophila let-7-Complex that encodes three evolutionary conserved and differentially expressed miRNAs (miR-100, let-7 and miR-125) and examined the expression and biological activity of the encoded miRNAs. The kinetic effects of Drosha and Dicer processing on the chimeric precursors were examined by in vitro processing assays. Our results highlight the importance of stem-base and terminal loop sequences in differential expression of polycistronic miRNAs and provide evidence that processing of a particular miRNA in a polycistronic transcript is in part determined by the kinetics of processing of adjacent miRNAs in the same cluster. Overall, this analysis provides specific guidelines for achieving differential expression of a particular miRNA in a cluster by structurally induced changes in primary miRNA (pri-miRNA) sequences.
MicroRNAs (miRNAs) belong to the class of small noncoding RNAs that regulate gene expression at the post-transcriptional level. Approximately 50% of miRNA loci in Drosophila melanogaster reside in close proximity in the genome to form clusters that are transcribed together. Since, these clusters are transcribed as a single primary transcript, fine-tuning of the downstream effector pathways is achieved by post-transcriptional regulatory mechanisms that determine the processing efficiency. The cis-acting sequence and structural features of the primary miRNA that interact with the processing machinery are critical determinants of the processing efficiency of the primary and precursor miRNAs. To elucidate the structural attributes of a polycistronic transcript that contribute towards the differences in efficiencies of processing of the co-transcribed miRNAs, we constructed a series of chimeric variants of Drosophila let-7-Complex that encodes three evolutionary conserved and differentially expressed miRNAs (miR-100, let-7 and miR-125) and examined the expression and biological activity of the encoded miRNAs. The kinetic effects of Drosha and Dicer processing on the chimeric precursors were examined by in vitro processing assays. Our results highlight the importance of stem-base and terminal loop sequences in differential expression of polycistronic miRNAs and provide evidence that processing of a particular miRNA in a polycistronic transcript is in part determined by the kinetics of processing of adjacent miRNAs in the same cluster. Overall, this analysis provides specific guidelines for achieving differential expression of a particular miRNA in a cluster by structurally induced changes in primary miRNA (pri-miRNA) sequences.
https://www.frontiersin.org/articles/10.3389/fcell.2022.909212/full

