Mismatch detection in A family DNA pol
- May 17, 2024
- Biotech Research, Biotechnology News, Publications, Recent Happenings, Research, Science News@RCB
- Poorti Kathpalia
- No Comments.
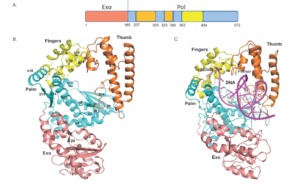
Figure: (A) Domain organization of ApPol1 (B) Crystal structure of ApPol1 and (C) Computational model of DNA-bound structure of ApPol1.
Replication of the genome is carried out high fidelity DNA polymerases in all organisms. The DNA polymerases are classified into seven families among which the A, B, C and D families are composed of high fidelity enzymes that are responsible for chromosome duplication.
In A-family DNA polymerases (dPols), a functional 3’-5’ exonuclease activity is known to proofread newly synthesized DNA. The identification of a mismatch in substrate DNA leads to transfer of the primer strand from the polymerase active site to the exonuclease active site.
To shed more light regarding the mechanism responsible for the detection of mismatches, we have utilized DNA polymerase 1 from Aquifex pyrophilus (ApPol1). Using a combination of structural and biochemical tools, we have identified a conserved residue in A-family DNA polymerases that plays an important role in mismatch detection. Also, our findings suggest that conserved residues lining the active site may detect mismatches at different positions in newly synthesized duplex DNA to enhance the fidelity of DNA synthesis.
Research Article: A conserved polar residue plays a critical role in mismatch detection in A-family DNA polymerases. Clement PC, Sapam T, Nair DT. Int J Biol Macromol. 2024 Jun;269(Pt 2):131965.